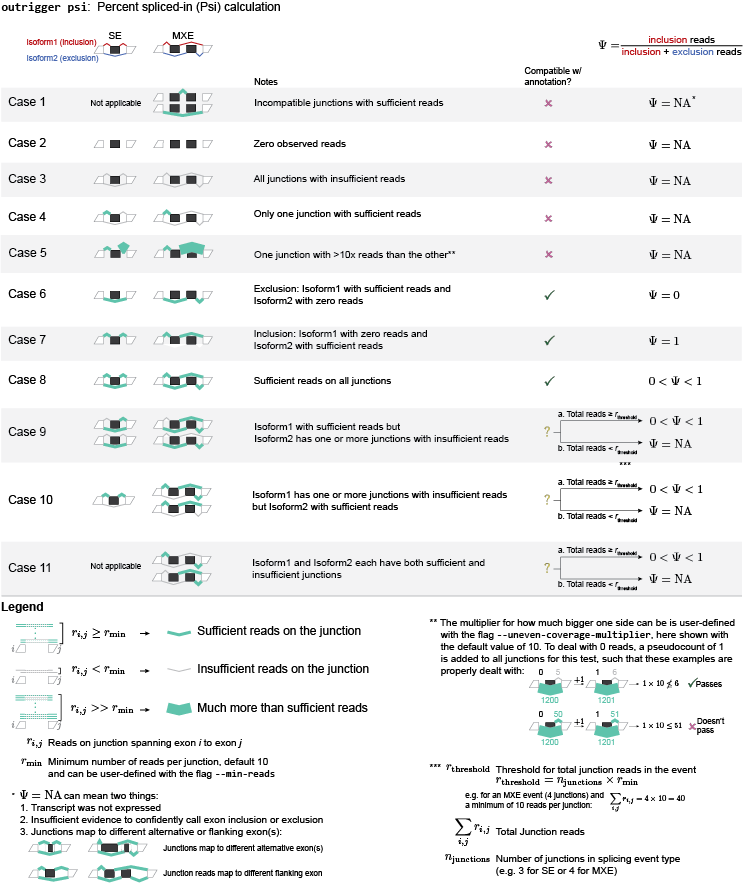
psi: Calculate percent spliced-in (Psi/Ψ) scores for your data from the splicing events you created¶
Overview¶
outrigger psi uses the splicing events created by outrigger index, or,
if outrigger validate has been run, only the valid alternative splicing
events with appropriate splice sites.
Inputs¶
If you used the default output folders for outrigger index and outrigger validate, then you can use the command,
outrigger psi
To run the percent spliced-in calculation.
The above command is equivalent to specifying all the arguments with their default values:
outrigger psi --index ./outrigger_index --min-reads 10
Below is the full usage output when you outrigger psi -h:
$ outrigger psi -h
usage: outrigger psi [-h] [-i INDEX] [-o OUTPUT]
[-c COMPILED_JUNCTION_READS | -j [SJ_OUT_TAB [SJ_OUT_TAB ...]]
| -b [BAMS [BAMS ...]]] [-m MIN_READS]
[--ignore-multimapping] [--reads-col READS_COL]
[--sample-id-col SAMPLE_ID_COL]
[--junction-id-col JUNCTION_ID_COL] [--debug]
[--n-jobs N_JOBS]
optional arguments:
-h, --help show this help message and exit
-i INDEX, --index INDEX
Name of the folder where you saved the output from
"outrigger index" (default is
./outrigger_output/index, which is relative to the
directory where you called this program, assuming you
have called "outrigger psi" in the same folder as you
called "outrigger index")
-o OUTPUT, --output OUTPUT
Name of the folder where you saved the output from
"outrigger index" (default is ./outrigger_output,
which is relative to the directory where you called
the program). Cannot specify both an --index and
--output with "psi"
-c COMPILED_JUNCTION_READS, --compiled-junction-reads COMPILED_JUNCTION_READS
Name of the compiled splice junction file to calculate
psi scores on. Default is the '--output' folder's
junctions/reads.csv file. Not required if you specify
SJ.out.tab files with '--sj-out-tab'
-j [SJ_OUT_TAB [SJ_OUT_TAB ...]], --sj-out-tab [SJ_OUT_TAB [SJ_OUT_TAB ...]]
SJ.out.tab files from STAR aligner output. Not
required if you specify a file with "--compiled-
junction-reads"
-b [BAMS [BAMS ...]], --bams [BAMS [BAMS ...]]
Bam files to use to calculate psi on
-m MIN_READS, --min-reads MIN_READS
Minimum number of reads per junction for calculating
Psi (default=10)
--ignore-multimapping
Applies to STAR SJ.out.tab files only. If this flag is
used, then do not include reads that mapped to
multiple locations in the genome, not uniquely to a
locus, in the read count for a junction. If inputting
"bam" files, then this means that reads with a mapping
quality (MAPQ) of less than 255 are considered
"multimapped." This is the same thing as what the STAR
aligner does. By default, this is off, and all reads
are used.
--reads-col READS_COL
Name of column in --splice-junction-csv containing
reads to use. (default='reads')
--sample-id-col SAMPLE_ID_COL
Name of column in --splice-junction-csv containing
sample ids to use. (default='sample_id')
--junction-id-col JUNCTION_ID_COL
Name of column in --splice-junction-csv containing the
ID of the junction to use. Must match exactly with the
junctions in the index.(default='junction_id')
--debug If given, print debugging logging information to
standard out
--n-jobs N_JOBS Number of threads to use when parallelizing psi
calculation and file reading. Default is -1, which
means to use as many threads as are available.
Outputs¶
Now the outrigger_output folder has psi subfolder, with the MXE
and SE events separate.
$ tree outrigger_output
outrigger_output
├── index
│ ├── gtf
│ │ ├── gencode.vM10.annotation.subset.gtf
│ │ ├── gencode.vM10.annotation.subset.gtf.db
│ │ └── novel_exons.gtf
│ ├── junction_exon_direction_triples.csv
│ ├── mxe
│ │ ├── event.bed
│ │ ├── events.csv
│ │ ├── exon1.bed
│ │ ├── exon2.bed
│ │ ├── exon3.bed
│ │ ├── exon4.bed
│ │ ├── intron.bed
│ │ ├── splice_sites.csv
│ │ └── validated
│ │ └── events.csv
│ └── se
│ ├── event.bed
│ ├── events.csv
│ ├── exon1.bed
│ ├── exon2.bed
│ ├── exon3.bed
│ ├── intron.bed
│ ├── splice_sites.csv
│ └── validated
│ └── events.csv
├── junctions
│ ├── metadata.csv
│ └── reads.csv
└── psi
├── mxe
│ └── psi.csv
├── outrigger_psi.csv
└── se
└── psi.csv
10 directories, 26 files