Fast detection and accurate calculation of alternative splicing with outrigger¶
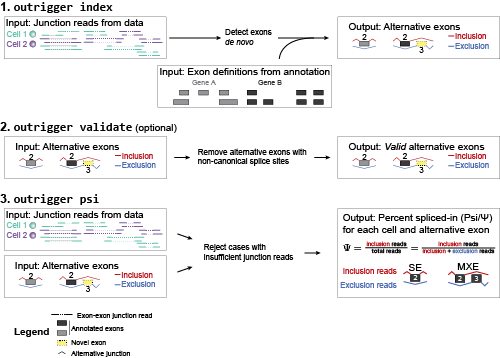
outrigger is a program which uses junction reads from RNA seq data, and a graph database to create a de novo alternative splicing annotation with a graph database, and quantify percent spliced-in (Psi) of the events.
- Free software: BSD license
- Documentation is available here: http://yeolab.github.io/outrigger/
Features¶
- Finds novel splicing events, including novel exons!
(
outrigger index) from.bamfiles - (optional) Validates that exons have correct splice sites, e.g. GT/AG
and AT/AC for mammalian systems (
outrigger validate) - Calculate “percent spliced-in” (Psi/Ψ) scores for all your samples
given the validated events (or the original events if you opted not
to validate) via
outrigger psi
Installation¶
To install outrigger, we recommend using the Anaconda Python
Distribution and creating an environment.
You’ll want to add the bioconda channel to make installing bedtools and its
Python wrapper, pybedtools easy, as these programs are necessary for both
outrigger index and outrigger validate.
conda config --add channels r
conda config --add channels bioconda
Create an environment called outrigger-env. Python 2.7, Python 3.4,
Python 3.5, and Python 3.6 are supported.
conda create --name outrigger-env outrigger
Now activate that environment:
source activate outrigger-env
To check that it installed properly, try the command with the help
option (-h), outrigger -h. The output should look like this:
$ outrigger -h
usage: outrigger [-h] [--version] {index,validate,psi} ...
outrigger (1.0.0dev). Calculate "percent-spliced in" (Psi) scores of
alternative splicing on a *de novo*, custom-built splicing index -- just for
you!
positional arguments:
{index,validate,psi} Sub-commands
index Build an index of splicing events using a graph
database on your junction reads and an annotation
validate Ensure that the splicing events found all have the
correct splice sites
psi Calculate "percent spliced-in" (Psi) values using the
splicing event index built with "outrigger index"
optional arguments:
-h, --help show this help message and exit
--version show program's version number and exit
Bleeding edge code from Github (here)¶
For advanced users, if you have git and Anaconda Python installed, you can:
- Clone this repository
- Change into that directory
- Create an environment named
outrigger-envwith the necessary packages from Anaconda and the Python Package Index (PyPI). - Activate the environment
These steps are shown in code below.
git clone https://github.com/YeoLab/outrigger.git
cd outrigger
conda env create --file environment.yml
source activate outrigger-env
Quick start¶
If you just want to know how to run this on your data with the default
parameters, start here. Let’s say you performed your alignment in the
folder called ~/projects/tasic2016/analysis/tasic2016_v1, and that’s
where your SJ.out.tab files from the STAR aligner are (they’re
output into the same folder as the .bam files). First you’ll need to
change directories to that folder with cd.
cd ~/projects/tasic2016/analysis/tasic2016_v1
Then you need find all alternative splicing events, which you do by
running outrigger index on the splice junction files and the gtf.
Note
We highly recommend to use outrigger index on a supercomputer with
multiple processors (at least 4) as the indexing process takes a long time
– over 24 hours, and by using multiple threads, the program will run much
faster.
Here is an example command:
Input: .SJ.out.tab files (faster)¶
This is faster than using .bam files because the junction counts are
already aggregated.
outrigger index --sj-out-tab *SJ.out.tab \
--gtf /projects/ps-yeolab/genomes/mm10/gencode/m10/gencode.vM10.annotation.gtf
Input: .bam files (slower)¶
If you’re using .bam files instead of SJ.out.tab files, never despair!
This will be slightly slower because outrigger needs to count every time a read
spans an exon-exon junction.
Below is an example command. Keep in mind that for this program to work, the
bams must be sorted and indexed.
outrigger index --bam *sorted.bam \
--gtf /projects/ps-yeolab/genomes/mm10/gencode/m10/gencode.vM10.annotation.gtf
Next, you’ll want to validate that the splicing events you found follow
biological rules, such as being containing GT/AG (mammalian major
spliceosome) or AT/AC (mammalian minor splicesome) sequences. To do
that, you’ll need to provide the genome name (e.g. mm10) and the
genome sequences. An example command is below:
outrigger validate --genome mm10 \
--fasta /projects/ps-yeolab/genomes/mm10/GRCm38.primary_assembly.genome.fa
Finally, you can calculate percent spliced in (Psi) of your splicing events! Thankfully this is very easy:
outrigger psi
It should be noted that ALL of these commands should be performed in the same directory, so no moving.
Quick start summary¶
Here is a summary the commands in the order you would use them for outrigger!
cd ~/projects/tasic2016/analysis/tasic2016_v1
outrigger index --sj-out-tab *SJ.out.tab \
--gtf /projects/ps-yeolab/genomes/mm10/gencode/m10/gencode.vM10.annotation.gtf
outrigger validate --genome mm10 \
--fasta /projects/ps-yeolab/genomes/mm10/GRCm38.primary_assembly.genome.fa
outrigger psi
This will create a folder called outrigger_output, which at the end
should look like the one below. Each file and folder is annotated with which command
produced it.
$ tree outrigger_output
outrigger_output...................................................index
├── index..........................................................index
│ ├── gtf........................................................index
│ │ ├── gencode.vM10.annotation.gtf............................index
│ │ ├── gencode.vM10.annotation.gtf.db.........................index
│ │ └── novel_exons.gtf........................................index
│ ├── exon_direction_junction_triples.csv........................index
│ ├── mxe........................................................index
│ │ ├── event.bed..............................................index
│ │ ├── events.csv.............................................index
│ │ ├── exon1.bed..............................................index
│ │ ├── exon2.bed..............................................index
│ │ ├── exon3.bed..............................................index
│ │ ├── exon4.bed..............................................index
│ │ ├── intron.bed.............................................index
│ │ ├── splice_sites.csv....................................validate
│ │ └── validated...........................................validate
│ │ └── events.csv......................................validate
│ └── se.........................................................index
│ ├── event.bed..............................................index
│ ├── events.csv.............................................index
│ ├── exon1.bed..............................................index
│ ├── exon2.bed..............................................index
│ ├── exon3.bed..............................................index
│ ├── intron.bed.............................................index
│ ├── splice_sites.csv....................................validate
│ └── validated...........................................validate
│ └── events.csv......................................validate
├── junctions......................................................index
│ ├── metadata.csv...............................................index
│ └── reads.csv..................................................index
└── psi..............................................................psi
├── mxe..........................................................psi
| ├── psi.csv..................................................psi
│ └── summary.csv..............................................psi
├── outrigger_psi.csv............................................psi
└── se...........................................................psi
├── psi.csv..................................................psi
└── summary.csv..............................................psi
10 directories, 26 files
Approximate runtimes¶
- Here are the expected runtimes for the different steps of
outrigger. In all - cases, we strongly recommend using a supercomputer with at least 4 cores, ideally 8-16.
outrigger index: This will run for 24-48 hours.outrigger validate: This will take 2-4 hours.outrigger psi: This will run for 4-8 hours.
Contents:
- Installation
- Usage
- Contributing
- Credits
- History
- v1.0.0 (April 3rd, 2017)
- v0.2.9 (November 11th, 2016)
- v0.2.8 (October 23rd, 2016)
- v0.2.7 (October 23rd, 2016)
- v0.2.6 (September 15th, 2016)
- v0.2.5 (September 14th, 2016)
- v0.2.4 (September 14th, 2016)
- v0.2.3 (September 13th, 2016)
- v0.2.2 (September 12th, 2016)
- v0.2.1 (September 12th, 2016)
- v0.2.0 (September 9th, 2016)
- v0.1.0 (May 25, 2016)